Affinity prediction using a neural network model for the m1 protein of group a streptococcus in customized medicine
Keywords:
Neural Network, Drug Affinity, M1 Protein, GAS Infection, Deep Neural Network.Abstract
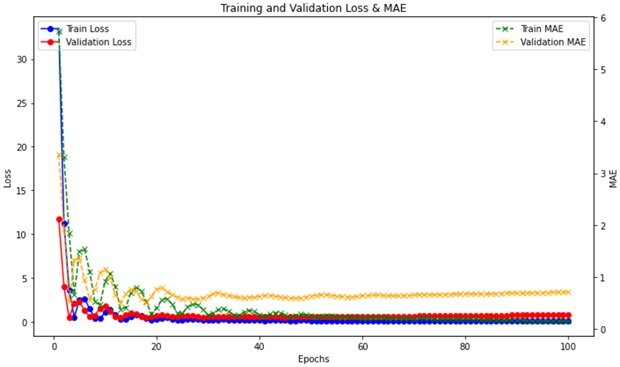
The M1 protein of Group A Streptococcus (GAS) is a critical virulence factor contributing to a wide range of diseases, from common pharyngitis to life-threatening invasive infections and autoimmune sequelae. Its role in immune evasion, inflammation, and tissue invasion makes it an attractive target for drug and vaccine development. Addressing M1-mediated pathogenesis is crucial to reducing the impact of GAS infections, particularly in vulnerable populations like children. This research study proposes the dry lab approach that leverages computational methods to provide valuable pre-analysis insights to optimise the wet lab experiments. This approach aims to reduce the experimental costs and time by predicting key outcomes in silico, guiding researchers in selecting the most promising candidates for further validation in the wet lab. The affinity of drug-protein interaction is central to developing effective treatments against M1 protein-associated GAS infections. High-affinity drugs can inhibit M1 functions, reduce immune-mediated damage, and ensure specificity and safety. By targeting M1 with high precision, these drugs can significantly mitigate the global burden of GAS diseases. This research study shows that using neural networks to predict drug-protein affinity to know the pros and cons of the proposed method that can significantly accelerate and enhance the development of therapeutics targeting the M1 protein of Group A Streptococcus (GAS).
Published
How to Cite
Issue
Section
Copyright (c) 2025 Chaitra Gajulla Nagaraja, Dr. Daisy Vanitha John, Pratham Tarachand Lalwani

This work is licensed under a Creative Commons Attribution 4.0 International License.




